Targeting Ribonucleases with Small Molecules and Bifunctional Molecules
Ribonucleases (RNases) cleave and process RNAs, thereby regulating biogenesis, metabolism and degradation of coding & noncoding RNAs. Hence, small molecules targeting RNases have the potential to perturb RNA biology, therefore RNases have been studied as therapeutic targets of antibiotics, antivirals and also agents for several cancers and autoimmune diseases. Additionally, recent advances in the chemically induced proximity approaches have led to the discovery of bifunctional molecules that target RNases to achieve RNA degradation or inhibit RNA processing. This article summarizes the efforts that have been made to discover small-molecule inhibitors and activators targeting bacterial, viral and human RNases. Also, the emerging examples of RNase-targeting bifunctional molecules and trends in developing such molecules for both the biological and therapeutic applications have been highlighted.
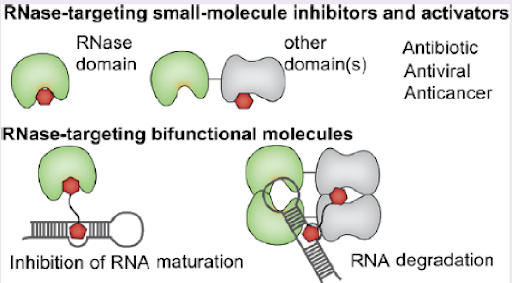
In humans, RNases belong to a superfamily constituted of at least 122 proteins that not only act as simple RNA degraders but are also involved in essential non-digestive cellular functions required in immune response, RNA maturation and angiogenesis. Distinction of these processes relies on the differences in RNA-substrate sequence of RNases and specificity for single- or double-stranded RNAs. RNases either function in a processive manner or cleave their substrate only once after binding.
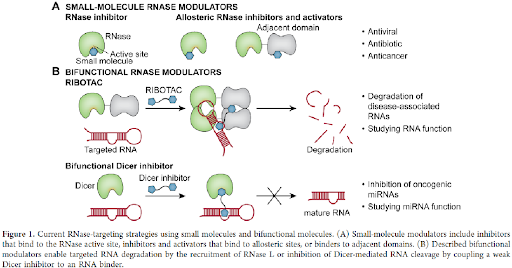
The nucleases are master regulators of RNA-dependent pathways and play indispensable roles in RNA biogenesis. Therefore, small molecules modulating RNase activities are useful tools for studying the regulatory mechanisms involving both human and pathogenic RNases and are potential candidates for the development of therapeutics. Representative examples of such small molecules include inhibitors of viral RNases influenza polymerase acidic (PA) endonuclease, human immunodeficiency virus (HIV) RNase H, and several human RNases. The potential of RNases as therapeutic targets is being discussed here summarizing the examples of these RNase-targeting small molecules. To note, in addition to small molecules, RNases have been targeted by the emerging class of proximity-inducing bifunctional molecules for various chemical biology applications. Therefore, in this review, two aspects of the current landscape in RNase targeting are highlighted: small molecules for the development of therapeutics and bifunctional molecules that modulate RNA biogenesis and metabolism (Figure 1). The former aspect is further divided into two target families, bacterial RNases and viral RNases.
SMALL-MOLECULE INHIBITORS OF BACTERIAL RNASES: Bacterial ribonuclease P (RNase P) is a unique RNase since it is a ribonucleoprotein functioning as a ribozyme. RNase P catalyzes the maturation of tRNA by cleavage of the 5′ end of precursor tRNA. Inhibitors of RNase P enzyme function, which is crucial for bacterial survival, could potentially serve as antibiotics. The only FDA-approved inhibitors of RNase P to date are antibiotic aminoglycosides such as Neomycin B. To note, RNase P inhibition is, however, not the sole mechanism of action for aminoglycoside antibiotics since their interaction with rRNA is known to cause errors in translation.
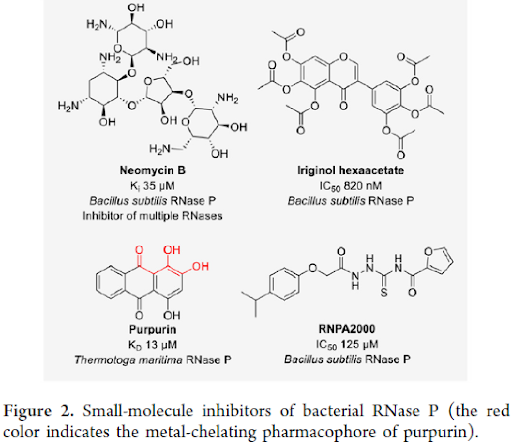
However, a screening of 2880 compounds discovered iriginol hexaacetate that inhibited RNase P with an IC50 of 0.8 μM. Similarly, Purpurin bound to RNase P with a KD of 13 μM and its binding to the protein component of the enzyme was confirmed by crystallization of a purpurin-RNase P complex. Purpurin has a three-oxygen pharmacophore that was often observed to coordinate the divalent metal ions of RNases. Additionally, RNPA2000 was reported to show weak RNase P inhibitory activity (IC50: 125 μM, Figure 2). A concern for the identified RNase P inhibitors, including RNPA2000, iriginol hexaacetate, and purpurin, was that they acted as aggregators and are likely unspecific RNase P inhibitors. In addition to small molecules, a FRET assay was employed to evaluate the rationally designed and modified oligonucleotides as inhibitors of RNase P. Antisense oligonucleotides targeting the RNA component of RNase P were coupled to cell-penetrating peptides to yield conjugates that inhibited bacterial growth.
SMALL-MOLECULE INHIBITORS TARGETING VIRAL RNASES: Small molecules have been reported for viral ribonucleases as antiviral therapeutics, such as those targeting HIV and HBV RNase H (Figure 3), influenza virus PA endonuclease (Figure 4), and the RNases nsp-14-ExoN and nsp15 of SARS-CoV-2 (Figure 4). RNase H is part of viral reverse transcriptase that removes RNA template strands to allow the synthesis of double-stranded DNA from viral genomic RNA, leading to the incorporation of the double-stranded DNA into the host genome by integrases.
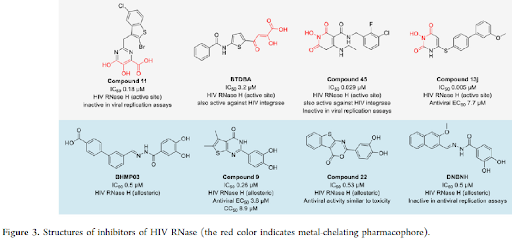
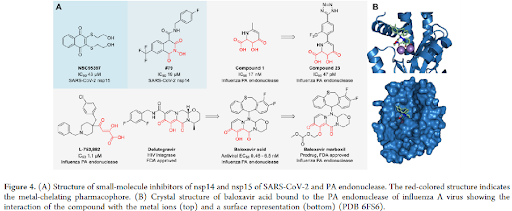
BIFUNCTIONAL MOLECULES TARGETING VIRAL RNASES: In general, the primary type of proximity inducing bifunctional molecules is proteolysis targeting chimeras (PROTACs) that have gained substantial attention in the past decade to achieve degradation of protein targets of interest via ubiquitination and proteasome-mediate degradation pathway. Until now, no PROTAC that degrades an RNase has been developed. A screening hit APL-16-5 against influenza PA endonuclease was confirmed to have a PROTAC-like mechanism. APL-16-5 is a microbial metabolite of Aspergillus sp. CPCC 400735 with an EC50 of 280 nM in antiviral assays that was shown to prevent the lethality. General structures and representative examples of Dicer-targeting bifunctional molecules and RNA-targeting RIBOTACs, instead of all reported examples with full structures, were shown (figure 7).
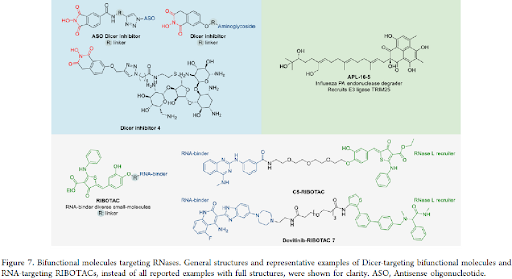
In comparison to the protein-degrading PROTACs, proximity-inducing bifunctional molecules have been reported to induce targeted degradation of RNAs via the recruitment of RNase L by ribonuclease targeting chimeras (RIBOTACs).
Concerning the biological relevance of RNases and their close associations with pathological states, it is reasonable to expect that more RNases will be subject both as the protein targets of interest for bifunctional molecules and as the effector protein components to be utilized by bifunctional molecules. These RNase-targeting molecules will contribute to probe an improved understanding of RNase biology at both the transcriptional and translational levels and provide new perspectives in developing small-molecule-based therapeutics.
- 1. Peng, Wu et al. Targeting Ribonucleases with Small Molecules and Bifunctional Molecules. ACS Chem. Biol. 2023, 18, 2101-2113.
- 2. Canestrari, E. et al. Ribonucleases as Drug Targets. Trends Pharmacol. Sci. 2018, 39, 855-866.
- 3. Schencking, I. et al. RNase P Inhibitors Identified as Aggregators. Antimicrob. Agents Chemother. 2021, 65, No. e00300-21.
- 4. Saramago, M.; et al. The nsp15 Nuclease as a Good Target to Combat SARS-CoV-2: Mechanism of Action and Its Inactivation with FDA-Approved Drugs. Microorganisms 2022, 10, 342.
- 5. Ren, Y. et al. Targeted inhibition of the endonuclease activity of influenza polymerase acidic proteins. Future Med. Chem. 2022, 14, 571−586.
- 6. Credille, C. V. et al. SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease. J. Med. Chem. 2019, 62, 9438−9449.
- 7. Li, G. et al. Approved HIV reverse transcriptase inhibitors in the past decade. Acta Pharm. Sin. B 2022, 12, 1567−1590.