Advances in RNA technologies
The basics:
RNA considered as a genetic middle man was probably evolved billions of years ago, in the earliest forms of life, even before the DNA was evolved. There are some major differences between DNA, the genetic material and RNA: (a) DNA is double stranded while RNA is a single strand (b) DNA backbone sugar is deoxyribose while the RNA consists of ribose (c) While DNA is present in every cell consistently, RNA is produced on need basis. The nitrogenous bases in RNA are adenine, guanine and cytosine and uracil (instead of thymine which present in DNA). RNA is highly prone to hydrolysis because of a reactive –OH group. RNA structure was first described in 1965 by R.W. Holley. Three of the most studied RNAs are mRNA (messenger RNA), tRNA (transfer RNA) and rRNA (ribosomal RNA). These RNAs govern many regulatory processes in both normal and disease conditions. Messenger RNA or mRNA carries the information from DNA in the nucleus to the cytoplasm where proteins are translated. rRNAs encode the ribosomal proteins and importantly “read” the code from mRNA to initiate protein synthesis. tRNAs, smallest of RNAs bring specified amino acids to ribosome to assemble them into proteins. RNAs can also be divided as coding (cRNA) and noncoding (ncRNA) RNAs. ncRNAs can either be housekeeping or regulatory and are further classified based on their size as long (lncRNA) or small ncRNA. There are various ncRNAs such as microRNA (miRNA), small nuclear RNA (snRNA), small- interfering RNA (siRNA), circular RNA and so on. There is an abundance of literature on the emerging important biological roles of these ncRNAs. In cancer, for e.g both cancer promoting and tumor suppressive roles have been identified for both lncRNAs as well as miRNAs.

In this article the main focus will be on therapeutic approaches with RNAs, specifically mRNA and tRNA. Although mRNA was first reported in 1950s, it was first isolated in 1962 by Sydney Brenner et al. (For those who are interested, the actual story of how mRNA was discovered is beautifully explained by Matthew Cobb in an essay published in Current Biology Magazine, 2015). Although it was one of the breakthrough discoveries, it is interesting to note that it was never considered for a Nobel prize!
RNA as a therapeutic agent
mRNA vaccines
One of the important cellular responses that perhaps is a self defence mechanism, is destruction of any RNAs found outside the cell by RNAses. mRNAs also form an integral regulatory platform for the cell to turn the protein synthesis ‘on’ and ‘off’. These properties along with the short half-life have made mRNA highly suitable for use in vaccine approaches. The concept of making vaccines from mRNA was first materialized in 1978 while multiple earlier attempts failed largely because of the negative charge of mRNA which made them impermeable in to the cell! It was not until the COVID-19 pandemic, when the mRNA vaccines (Moderna’s Spikevax) saw the light of the day and turned out to be truly life-saving and a huge commercial success.
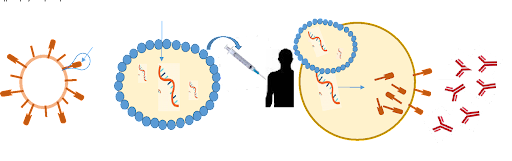
The idea behind the COVID-19 mRNA vaccine (Figure 1) is quite simple: Coronavirus has what is called as a ‘spike protein’ on their surface which helps them enter the human cells and the vaccine has a mRNA with instructions to make the spike protein. Once the mRNA is injected into the blood stream, the spike proteins are made that are recognized by the immune system as foreign to which immune response is triggered to destroy the spike protein. The vaccine’s mRNA then undergoes self-destruction as should be the case. This vaccine was indeed shown to be highly efficacious where there was a more than 94% reduction in the number of symptomatic COVID-19 cases among the 30,000 people who received the Spikevax vaccine as compared to those who received placebo injection.
But, there’s a catch here! Since mRNAs are destroyed by our immune system way too fast even before the immune response is triggered, scientists replaced one of the mRNA’s bases Uridine with ‘Pseudouridine’ so that they can evade self-destruction long enough to make the intended protein. This discovery indeed earned Katalin Karikó and Drew Weissman their Nobel prize for accelerating the development of mRNA based vaccine and therapy. But, according to a recent study from University of Cambridge, mRNAs with these ‘pseudouridines’ make significantly more frameshift proteins as compared to their uridine versions!! Just to clarify: all natural mRNAs sometimes get translated incorrectly and the question was if these altered mRNAs also get erroneously translated too! To test this, the scientists designed the mRNA to make a fluorescent protein when there’s a slip of a three- letter code (a frameshift); basically a fluorescent protein is produced if there’s a mistake in reading the codon!! This experiment indeed confirmed the production of many frameshift proteins, not just in laboratory, but also from immune responses observed in people vaccinated with Pfizer’s mRNA vaccine which was not observed in people who got Astrazeneca’s DNA vaccine! Essentially, it was confirmed beyond doubt that these mRNA vaccines do make many unintended proteins in the cells although as of now there’s nothing to indicate that these proteins are harmful!! And the good news is that scientists believe that these frameshifts can be avoided by alternating the ‘pseudouridines’ with a suitable base that codes for the same amino acid.
Nonetheless, the field of mRNA vaccines is marching ahead in full force with prophylactic vaccines being developed for respiratory diseases, cancer and some rare diseases. The limitations of mRNA vaccines so far are stability of the mRNA, identifying the right target protein, cellular delivery, protein expression from the mRNA vaccine in a cell and immunogenicity. All these parameters have to be taken into account and optimized to develop a successful therapeutic vaccine.
Treatment of rare diseases with tRNA
tRNAs are much smaller than mRNA and the largest tRNA comprises of only 76 nucleotides! In humans, there are about 47 types of tRNAs for decoding 20 standard amino acids plus selenocysteine. As mentioned earlier, each tRNA molecule reads a trio of nucleotides called a codon, (code for a particular amino acid) and accordingly transfers the particular amino acid to a protein.
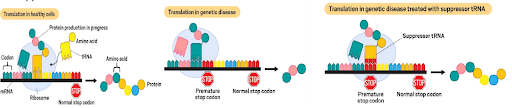
Due to what we call as a ‘non-sense’ mutation, a stop codon is produced in place of an amino acid and therefore, the protein produced is truncated with no or altered biological activity, which forms the basis for a disease called as “Stop Codon Disease” (SCD). This includes thousands of rare and common diseases and approximately 10% of people (~30 million worldwide) with genetic disease have SCD. In this disease, for e.g the code CGA codes for amino acid arginine and when CGA is mutated into UGA, it becomes a stop signal and the protein is no longer produced. In such cases suppressor tRNA, that recognizes the UGA codon and delivers arginine to continue the protein production, can be used as a therapeutic approach (Figure 2). Some of the dreaded diseases with high unmet medical need, such as cystic fibrosis, muscular dystrophy, Dravet’s syndrome, Rett syndrome etc. can all be tremendously benefitted by tRNA based therapeutics. Dravet’s disease for e.g is a rare type of epilepsy (with uncontrolled seizures) caused by stop codon mutation in a gene SCN1A that encodes a sodium channel. Similarly, 10% cystic fibrosis and two thirds of Duchene muscular dystrophy are caused by the stop codons and in such cases there can be tremendous benefit with use of tRNA based therapeutics.
Several start-up companies, ReCode Therapeutics, Shape Therapeutics, Tevard, AlltRNA and others now focus on therapeutic tRNAs and have raised millions of dollors. Alltrna recently demonstrated in an animal study, that their therapeutic tRNA can actually read these premature stop codons and deliver the right amino acid thereby producing the active full- length protein. Although it is still far from human use, it is certainly a big milestone in RNA therapeutics. But, challenges of drug development in terms of delivery and tolerability/toxicity still remain. We still need to thoroughly understand the selectivity of tRNA to the intended codon making sure that it does not cause a chaos to the protein translation machinery of the cell leading to toxicity. Although delivery of RNA therapeutics has always been challenging, the successful use of COVID-19 vaccines through nanoparticle based delivery system has opened up the potential for these RNA based therapies enhancing the hope of treating these dreaded diseases.
RNA modifying proteins as therapeutic targets
In addition to RNA being used as a treatment strategy, RNA modifying enzymes are also finding their place in RNA based therapeutics. RNA modifications play a critical role in several cellular process and changes in these modifications is associated with pathological conditions. There are more than hundred different post translational modifications reported that dynamically regulate RNA function and stability. Similar to DNA modifying proteins, RNA modifying proteins are also termed as ‘readers’, ‘writers’ and ‘erasers’.
Enzymes that add a methyl group on m6A are known as methyl transferases (METTL) and those that remove the methyl group are called as demethylases (ALKBH5). There are also m6A recognizing proteins called as ‘readers’ (YTHFD). In a normal cell, METTL3-mediated m6A methylation and dynamic interplay between ALKBH5 and METTL3 have been shown to be important for cardiomyocyte hypertrophic response, lysosome biogenesis and autophagy.
M6A dysregulation though has been reported to be associated with multiple pathological conditions and has been comparatively well studied in cancer. Recent studies on the m6A writer METTL3 or METTL14 indicate their oncogenic roles and METTL3 for example is abundantly expressed in Acute myelogenous leukemia (AML) and has been identified as a crucial gene for proliferation of AML. Accordingly, METTL3 depletion in human hematopoietic stem/progenitor cells (HSPCs) increased cell differentiation and reduced cell proliferation. Leukemic cells without METTL3 also failed to establish leukemia mouse xenograft and METTL3 has been increasingly recognized for its oncogenic role.
Similar to m6A, there are numerous other sites that undergo post transcriptional modifications and those include m1A, m7G, I, m6Am, Ψ, mcm5s2U, mcm5U, ac4C. In all these sites there’s dynamic interplay of writers, readers and erasers which are involved in multiple cellular processes. Key roles played by these proteins have been confirmed at the cellular level in multiple disease conditions, including metabolic diseases, cardiovascular diseases, genetic and developmental diseases, immune disorders, cancer and so on. But, therapeutics targeting these proteins is still its infancy with various companies putting together a lot of effort in this regard. We still need selective, potent inhibitors showing a benefit in animal studies or in clinical trials in any of the diseases mentioned above and further advanced studies are warranted in area to develop a targeted therapy.
- 1. Qiu et al., 2023 RNA modification: mechanisms and therapeutic targets. Review, Volume 4, article number 25, (2023)
- 2. Gretchen Vogel., mRNA vaccines may make unintended proteins, but there’s no evidence of harm: Alterations that help messenger RNA persist in living cells can trip up protein synthesis. doi: 10.1126/science.z60ty3k
- 3. https://www.science.org/content/article/mrna-vaccines-may-make-unintended- proteins-there-s-no-evidence-harm
- 4. Cross R., tRNA therapies could help restore proteins lost in translation: A new class of therapies based on transfer RNA could treat forms of cystic fibrosis, muscular dystrophy, genetic epilepsies, and more. Volume 99, Issue 34, 2021
- 5. https://www.labiotech.eu/podcast/beyond-biotech-podcast-66-treating-rare-diseases- with-trna/
- Author : Dr. Dhanalakshmi S Chief Scientific Officer at TheraIndx Lifesciences Private Limited